Nephele User Guide
Getting Started Using Nephele
Overview: Nephele is a cloud platform developed by a team of computational biologists and bioinformaticians that also perform metagenomics analysis in collaboration with researchers at NIAID. It was born from the need of making tools and pipelines available to those with limited computational resources or those lacking expertise in metagenomics pipeline development.
Before you use Nephele, you should know that the team has designed the user interface in a series of steps to help guide users through the order of steps recommended for analysis of most datasets. These steps are:
- Pre-process
- Analyze
- Explore
Step 1: Pre-process
We recommend that you always start by inspect the quality of your sequencing data and then proceed to pre-process it (e.g. trim, filter by quality, adapter removal).
Quality check pipelines are available for short read (Short Read QC and Oxford Nanopore sequence data (NanoporeQC)).
Step 1 will inspect and then prepare reads for input to the analysis pipelines. Even though certain analysis pipelines available on
Step 2 include a default quality filtering and even a pair merging step, users might prefer to run these ahead of time using this pipeline.
If you have already done a quality check of your data and are comfortable with the quality of your reads, proceed to Step 2 and start analyzing your data.
Step 2: Analyze
This step has a growing collection of compute-intensive pipelines that will generate tables of sequence variants, run metagenomics assemblies,
assign taxonomy and more. Ideally, the input data has already been examined using the tools available in the Step 1 QC pipeline;
remember "Garbage in, Garbage out". The output of some of the pipelines consist of tables and graphics but not everything ends there.
Users should download the output files, extract knowledge and also use some of those files (e.g. biom, fasta) as input for other
pipelines in the Step 3: Explore section.
Step 3: Explore
In this section users will find pipelines to extract further insights from the data after having completed the processing and analysis.
These pipelines will run various analyses and create publication quality graphics.
How to submit a job
Step 1: Select analysis type.
Please select the analysis you are interested in. Nephele provides amplicon analysis (16S, ITS), WGS metagenomics analysis and Viral (SARS-CoV-2 mapping).
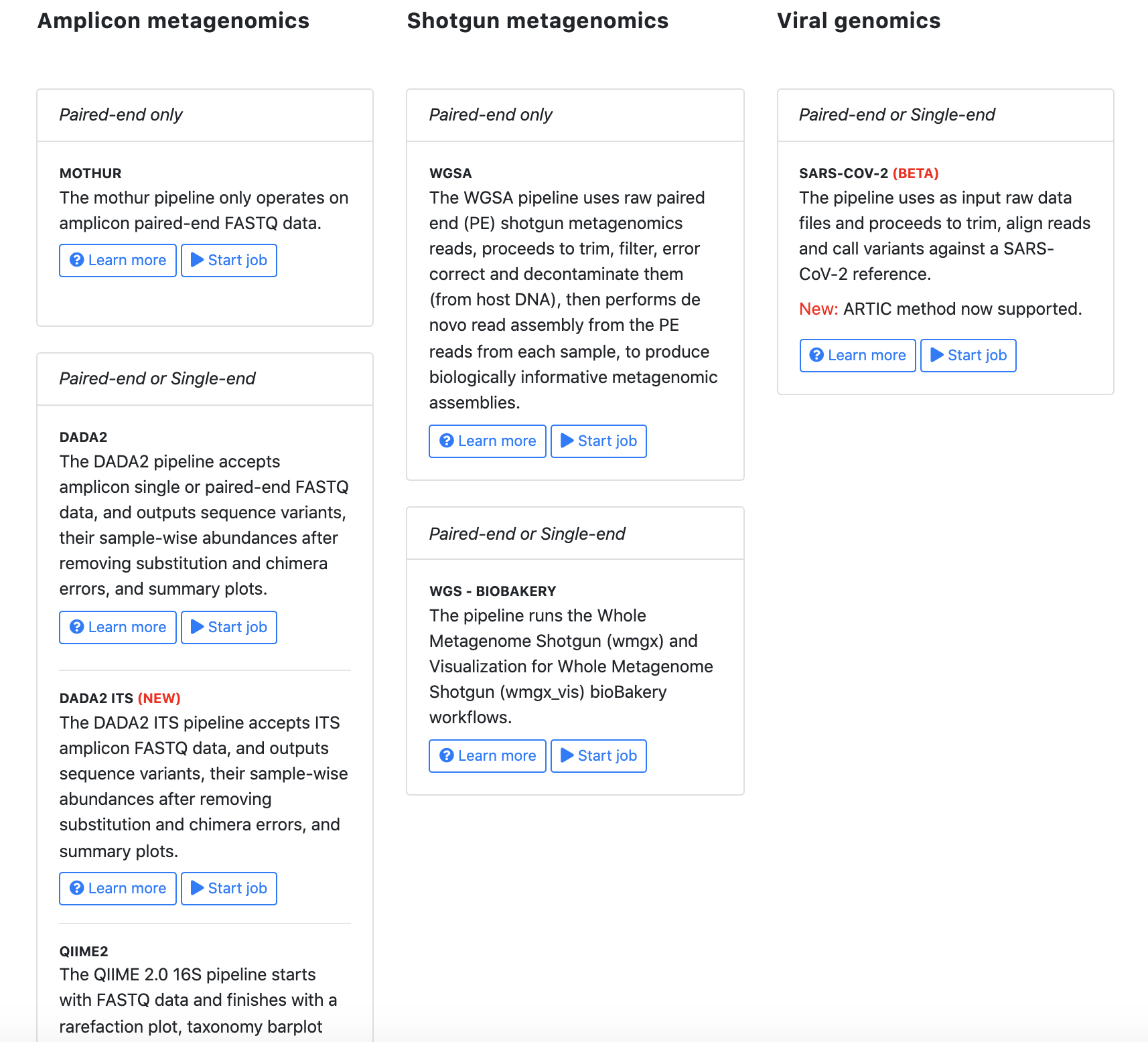
Step 2: Select data type (demultiplexed file only)
Nephele supports the following data types for analysis types.
- 16S
- demultiplexed paired end fastq (select this option if you are using the sample data)
- demultiplexed single end fastq
- ITS
- As ITS supports only demultiplexed paired end fastq, the system will assume the data type and proceed to next page
- WGS
- demultiplexed paired end fastq
- demultiplexed single end fastq
Select the type appropriate to your data (e.g. Paired End FASTQ) and click “Next”.
Step 3: Select upload method.
Nephele provides four methods to upload input files: my computer, Google Drive, BaseSpace Globus. Upload from my computer allows you to upload files from your local computer. This method is suitable for up to 2GB per fastq or fastq.gz file per file. We recommend that you upload compressed .gz files for performance. Select this option if you are using a sample dataset with small files. If you have a large data set, we recommend opting to use Globus, Google Drive or BaseSpace method.
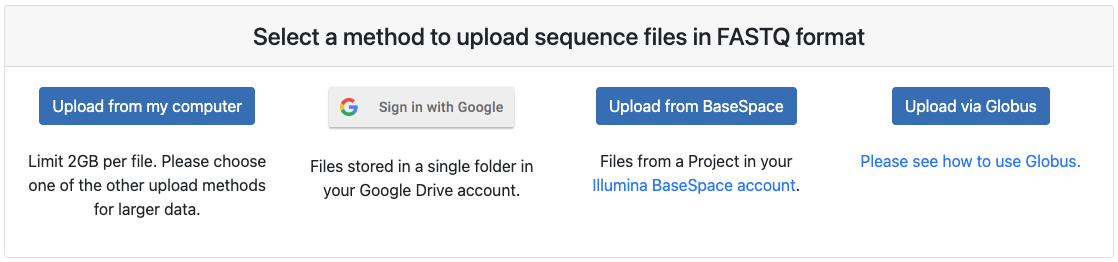
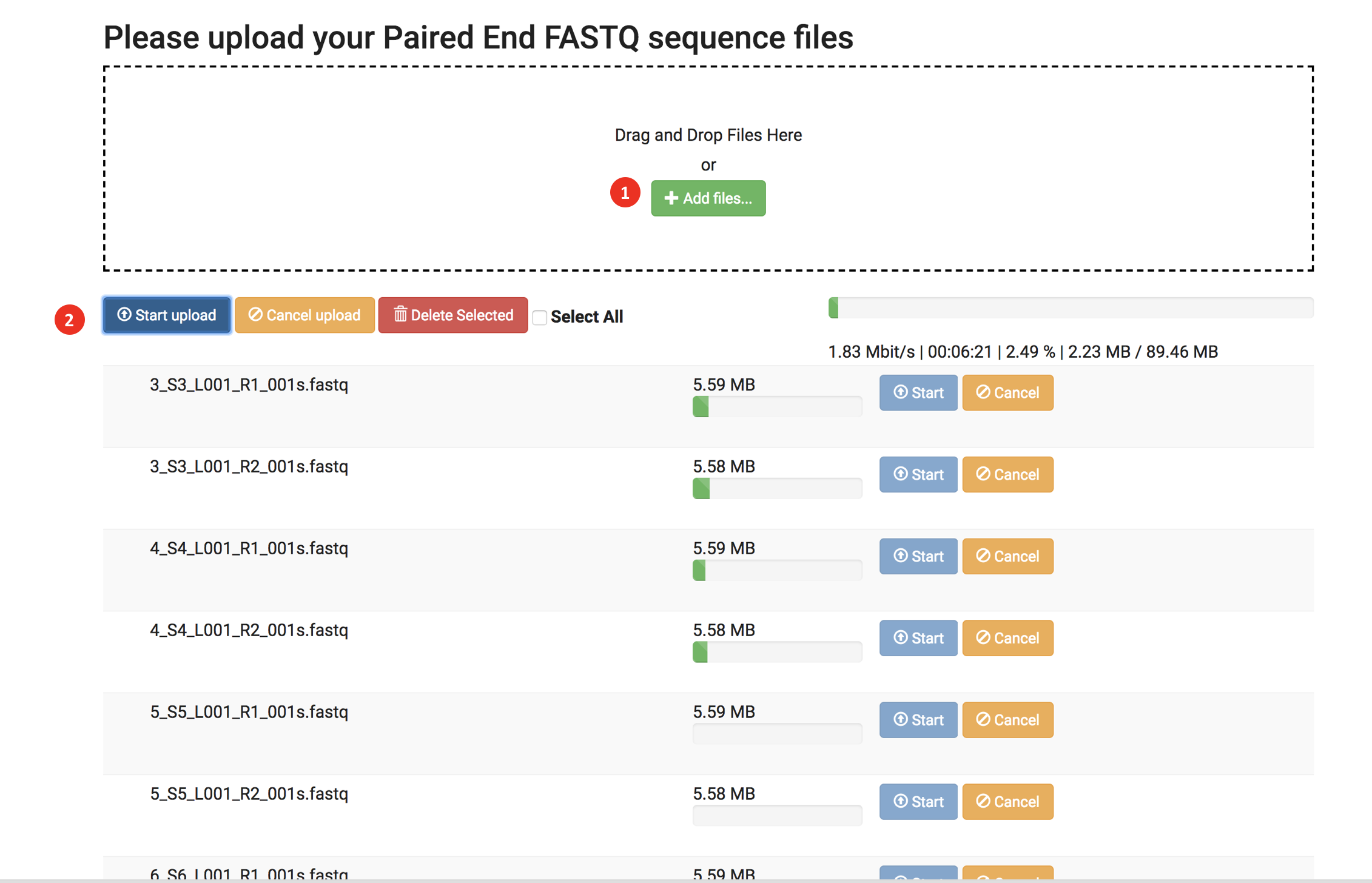
- Upload from my computer
- Click "+ Add files" and select fastq or fastq.gz files or Drag and drop the files.
- Click Start upload to start upload your input files.
*Dependent on your network, the upload speed and total time can vary. We recommend stable and high-speed internet for this method. - After completing the uploads, click Next.
- You can cancel the upload anytime by clicking the "Cancel upload" button.
- You can select all files by clicking "Select all."
- If you wish to delete files, you can delete them by clicking the "Delete selected" button.
- Google Drive: We recommend that you organize all of your input files in one folder ahead of time. During upload you will select the folder and all fastq files in that folder will be available for input. Proceed to the next step of uploading the mapping file. Nephele will proceed to import all the fastq files that correspond to the files indicated on the mapping file.
- BaseSpace: Nephele will display all available Projects in BaseSpace. During upload you will select the Project you wish to use, Nephele will find all the fastq files associated with that Project. Proceed to the next step of uploading the mapping file. Nephele will proceed to import all the fastq files that correspond to the files indicated on the mapping file.
- Globus (recommended for large data). Please see tutorial here.
Step 4: Upload mapping file
After uploading your input files, Nephele pipelines require a mapping file. To upload your mapping file, simply click Browse, select your mapping file, and click Upload. Upload the sample mapping file (xx.xlsx).
Nephele validates your mapping file instantly so that you can correct any error interactively. In order to prevent any mapping file errors, we recommend that you start with mapping file templates provided on the map file upload page for the pipeline you're running.
Step 4.1: How to use Nephele interactive mapping file validation
If you have an error in your mapping file, for example, a typo in your fastq file name column, here is an example of how to correct it using the interactive error page.
- On the validation error page, move your mouse over to the area that is highlighted in red.
- This example shows that the file names in both ForwardFastqFile and the ReverseFastqFile column names are the same: "3_S3_L001_R1_001s.fastq".
There is a typo on the ReverseFastqFile column.
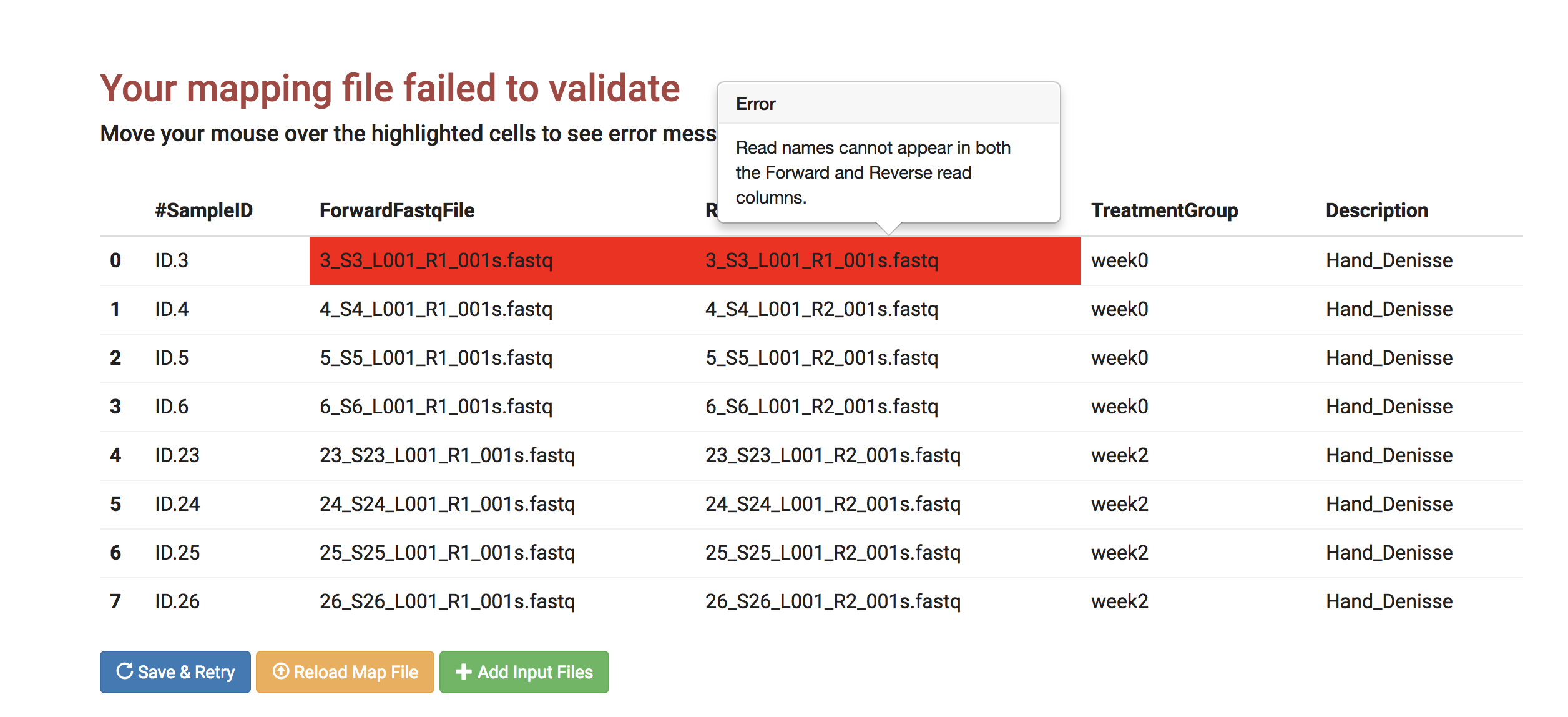
- Click on the ReverseFastqFile column and correct "3_S3_L001_R1_001s.fastq" to "3_S3_L001_R2_001s.fastq."
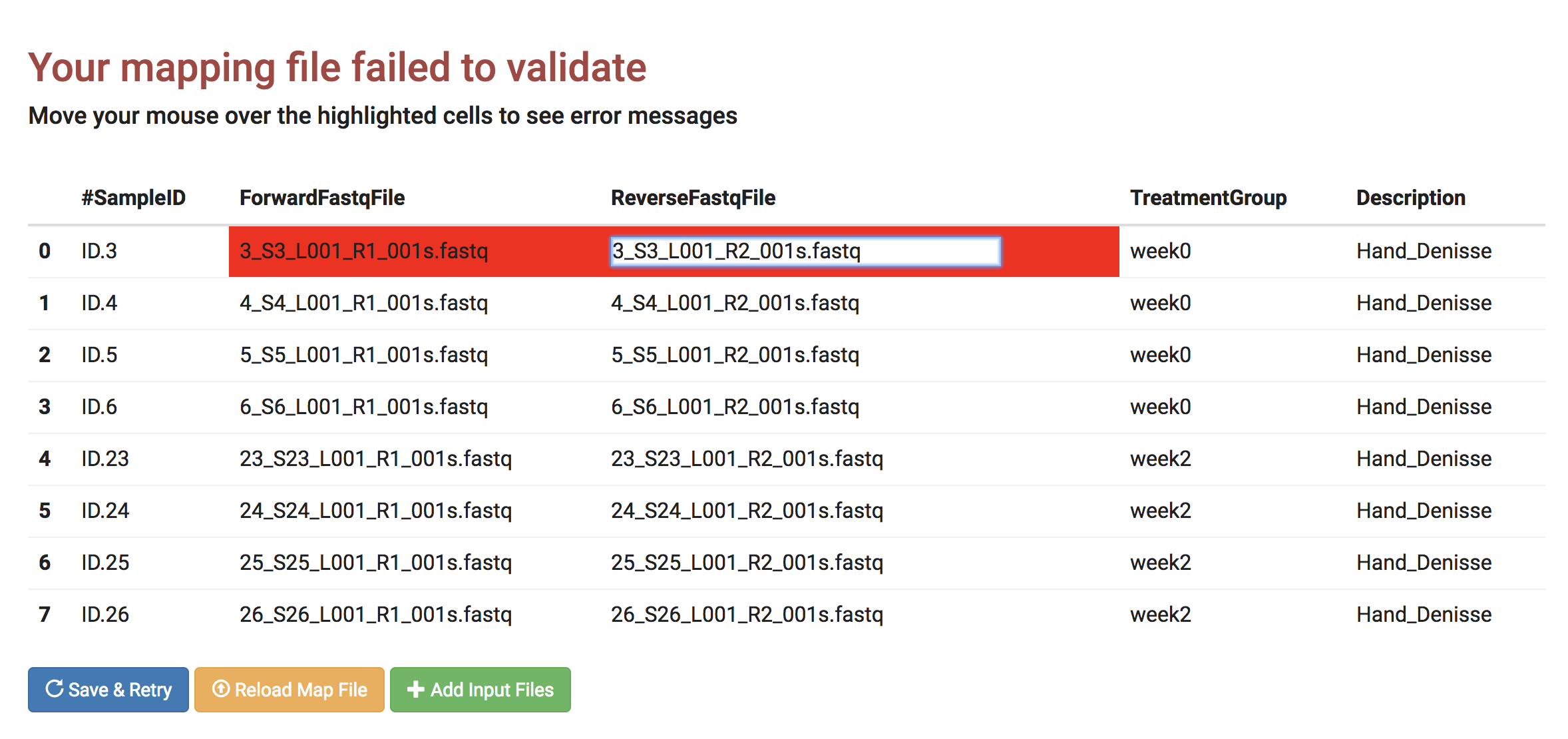
- Click "Save & Retry."
- The mapping file passes the validation and moves to the next page, Select Pipelines, automatically.
Notes on map file validation:
- "BarcodeSequence" and "LinkerPrimerSequence" columns are no longer required by Nephele pipelines. Please remove them if you are using older mapping files.
- The file names listed in the mapping file should exactly match the names of the files you have uploaded for analysis. If you have uploaded *.fastq.gz files, make sure to add the .gz extension to the file names in the mapping file. Do not include full file paths.
- Nephele 2 accepts tab-delimited text (.txt) files and Excel (.xlsx or .xls) files. Comma-separated (.csv) files are not supported.
Step 5: Update parameters (optional)
Once you select a pipeline, you will see a submission page. You can enter a description of your job so that you can identify different jobs easily.
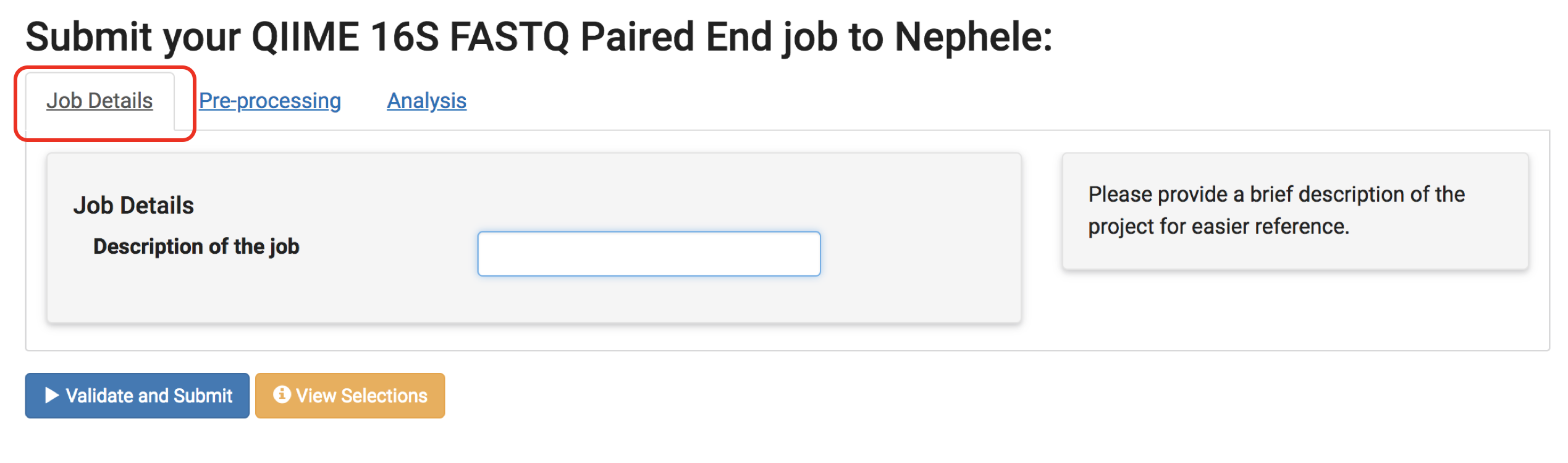
On this page, there are different tabs such as "Pre-processing" and "Analysis" (dependent on the pipeline) which allow you to change the parameters the pipeline is run with. Don’t forget to have a look if you are interested in different options. See Nephele's Pipeline Section for more details regarding parameters.
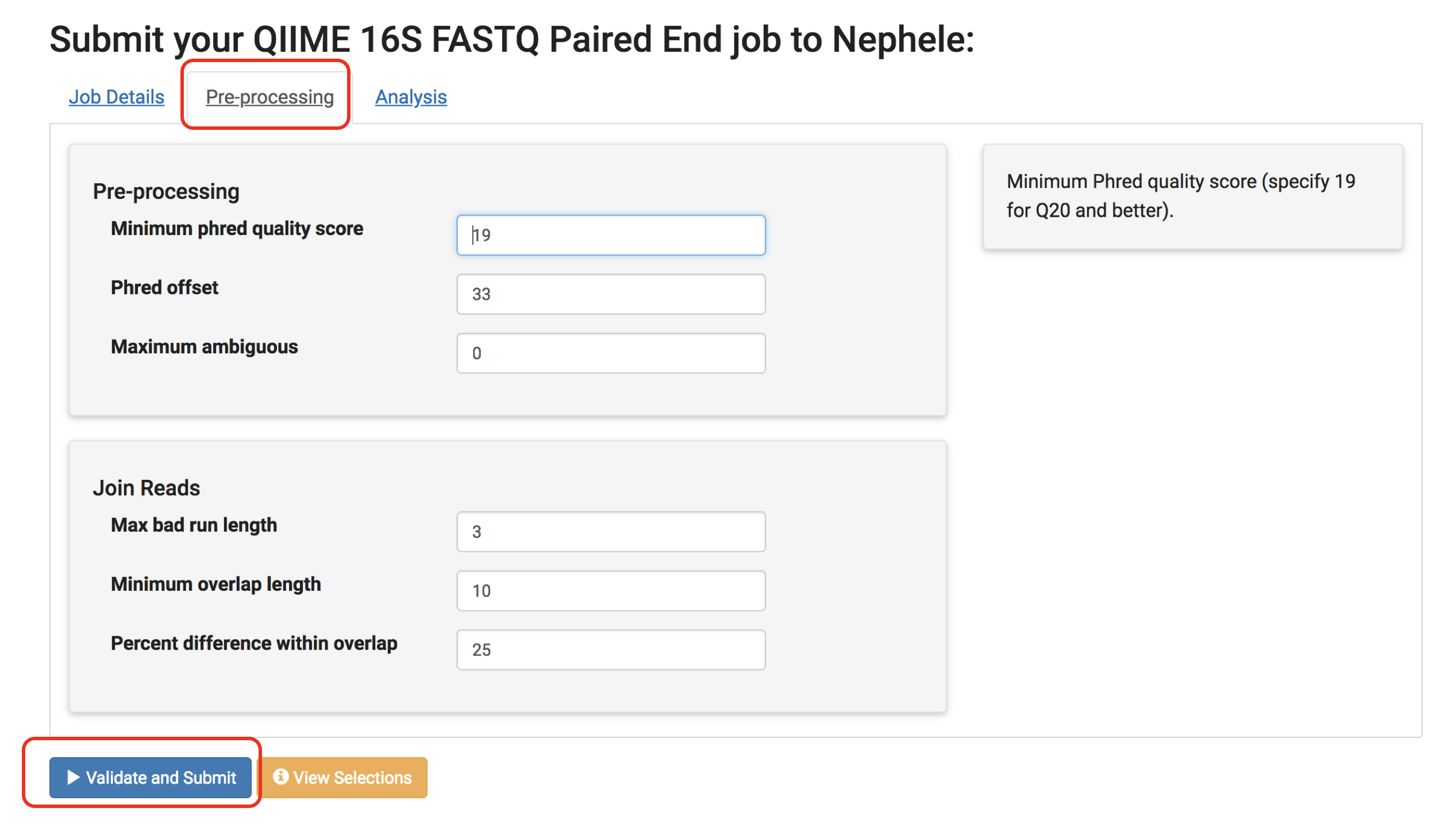
Finally, click Validate and Submit!
Congratulations! You have submitted a job! You will receive a Pipeline Started email shortly.
How to resubmit your job
You can resubmit a job without uploading all of your input files again.
Step 1: Enter the Nephele JobID that you would like to resubmit.
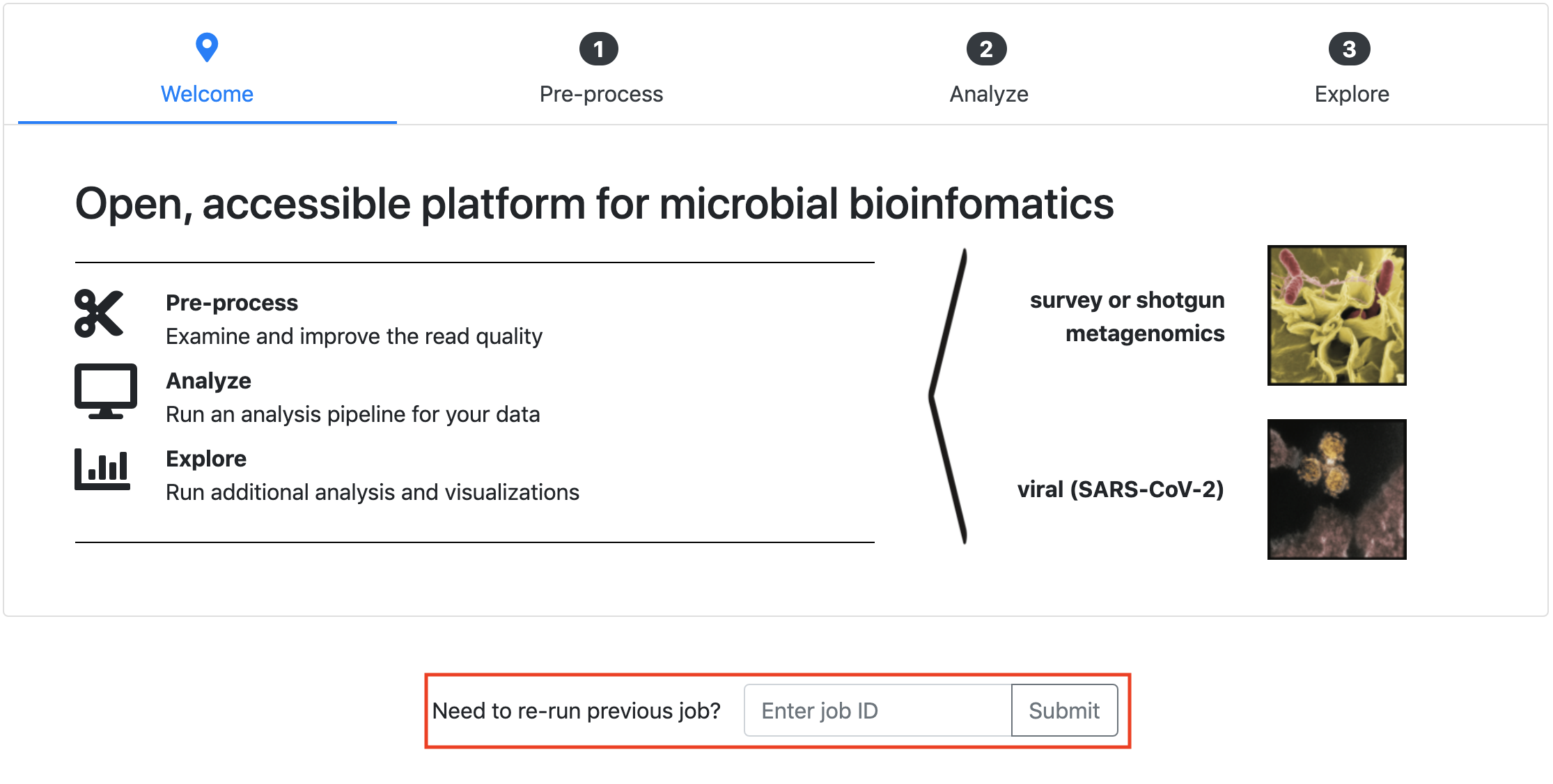
Step 2: As long as your job hasn't expired, your input data will be retrieved. The rest of steps are the same as submitting a job. You can re-upload a mapping file and change parameters for the resubmission.